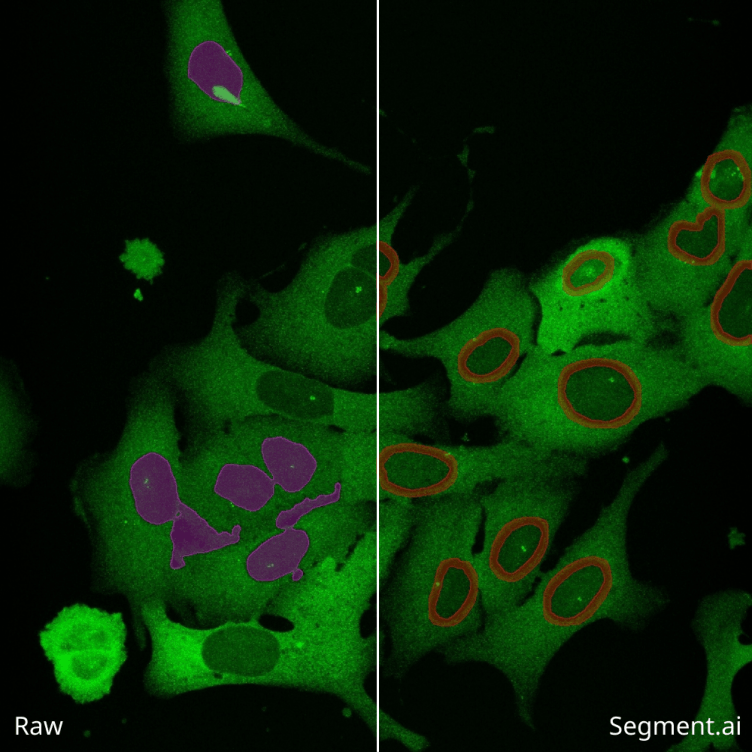
Excitation of a sample with Green Fluorescent Protein (GFP) dye
Principle
The principle of fluorescence in microscopy is based on the ability of certain molecules, called fluorophores, to absorb light at a specific wavelength and then emit light at a longer wavelength.
When a sample is fluorescently labeled, it means that specific molecules or structures of interest have been tagged with fluorescent dyes or proteins.